A novel computer model uses microorganisms’ DNA to determine the metabolites used and produced by the bacteria in different parts of the human body. Metabolites provide important clues about human health and disease but can be difficult to measure experimentally. The results of the research were published in Nature Microbiology.
Metabolites, such as those that make up one’s body odour, are molecules that are used and produced by living cells such as human cells and bacteria. The human body is increasingly seen as a complex ecosystem, home to thousands of microorganisms and their metabolites. The health of this ecosystem hinges on the relationship between the microbes and human cells and tissues, and metabolites are often the key to this relationship. For example, intestinal tumours cause changes to the metabolite composition in the intestine, which in turn allows certain types of gut bacteria to grow, sometimes exacerbating the tumour.
Difficult to determine
“Metabolites are vital, but it is often difficult to find out which ones are present at a specific site in the body. There are many different metabolites, and their concentrations can vary widely,” explains research leader Dr. Bas Dutilh from Utrecht University. “Since metabolites are so closely linked to the growth of bacteria, we thought that it might be possible to predict their composition based on the types of bacteria present. The only input we need for that is the abundances of the bacteria and their genes, and progress in the field of metagenomics has made it relatively easy to get those.”
Surprisinly consistent
Given gene information from the bacteria as input, the computer model simulates the most likely metabolites that the bacteria need to grow. These are found by checking which enzyme each gene encodes, and which metabolites each enzyme uses/produces. The researchers then calculate the concentrations of metabolites that explain the abundances of bacteria observed at a given site in the human body. “The predictions we arrive at in our analyses are surprisingly consistent with what is known about the metabolite concentrations in and on the human body,” explains lead author and PhD candidate Daniel Garza, affiliated with Radboudumc.
Applicable to any environment
“We have shown the utility of the computer model for the human skin, mouth, intestines and vagina, because those are fairly well-known areas,” says Prof. Martijn Huijnen from Radboudumc. “But in principle, this technique is applicable to any environment where bacteria live, such as the soil near plant roots.”
Beauty and skin care products
The researchers found that many of the substances predicted from skin samples are known to be ingredients of beauty and skin care products, such as myristic acid, a fragrance ingredient that is easily absorbed by the skin. The results also predicted a significant amount of lead on skin, which corresponds to the findings of a previous experimental study. Where the toxic metal comes from, however, is still anyone’s guess. Lead supplement in petrol has long been prohibited in the Netherlands and the United States, where the samples were taken. “It’s still used in some brands of lipstick, but that doesn’t seem to be a likely explanation for the relatively high concentrations found in so many skin samples”, according to Dutilh.
The model in detail
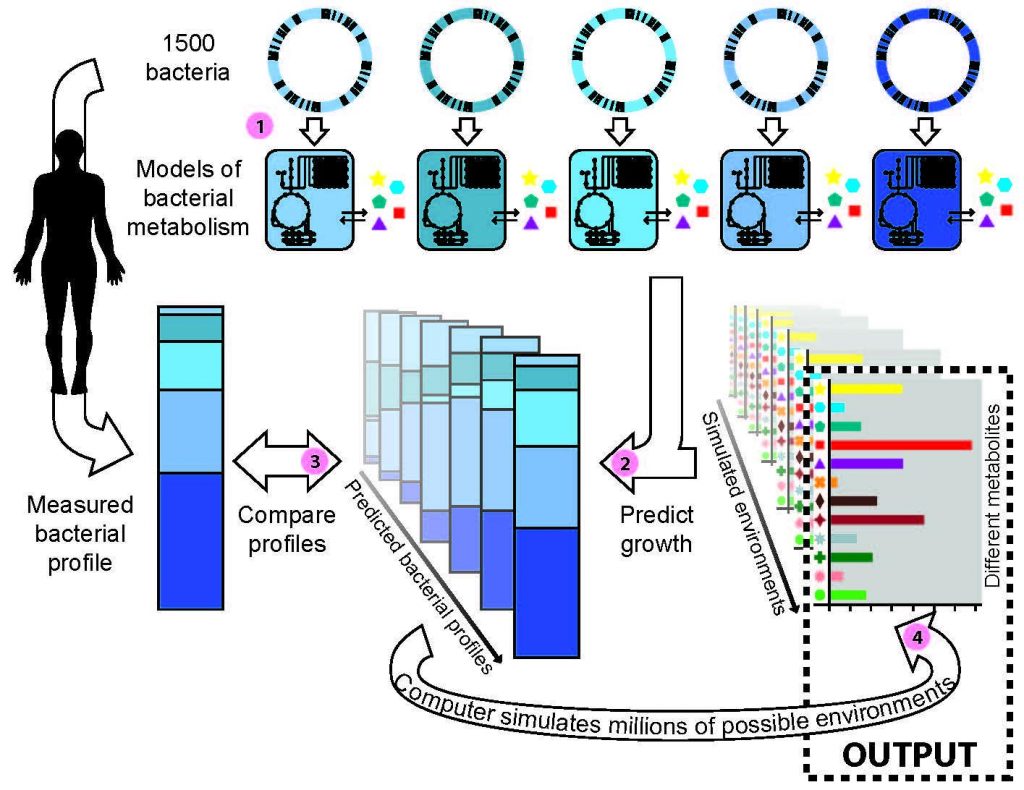
The computer algorithm, called MAMBO (for ‘Metabolomic Analysis of Metagenomes using fBa and Optimization’), works as follows (see figure). (1) First, computer models are created of the metabolisms of more than 1,500 bacteria from the human body. These models record the relationship between the concentration of metabolites in the environment and the growth of each bacteria. (2) The models are used to conduct computer simulations of bacterial growth on an initially random, but well-defined metabolite environment. This produces a predicted profile indicating which bacteria will grow in the selected metabolite environment, and how well they grow. (3) The predicted profile is then compared to an experimental profile, for example one listing the abundances of all the bacteria found in samples from skin or faeces. (4) After simulating millions and millions of environments, the predicted profile that best corresponds to the experimental profile is found. This prediction describes all the bacteria and metabolites that are found and at what concentrations. Thus, the metabolite concentrations in an environment can be predicted by measuring the bacterial genes.
Publication
‘Towards predicting the environmental metabolome from metagenomics with a mechanistic model‘
Daniel R. Garza, Marcel C. van Verk, Martijn A. Huynen and Bas E. Dutilh
Nature Microbiology, 12 March 2018, DOI 10.1038/s41564-018-0124-8
This research was funded in part by a VIDI grant awarded to Bas Dutilh by the Netherlands Organisation for Scientific Research (NWO), and by the Brazilian government programme ‘Science Without Borders’, which finances Daniel Garza’s PhD research in the Netherlands.