Research into the role of structural variants (SVs) in cancer has been limited due to difficulties in detection. UBC members Ianthe A. E. M. van Belzen, Alexander Schönhuth, Patrick Kemmeren & Jayne Y. Hehir-Kwa explore current strategies for integrating SV callsets and to enable the use of tumor-specific SVs in precision oncology.
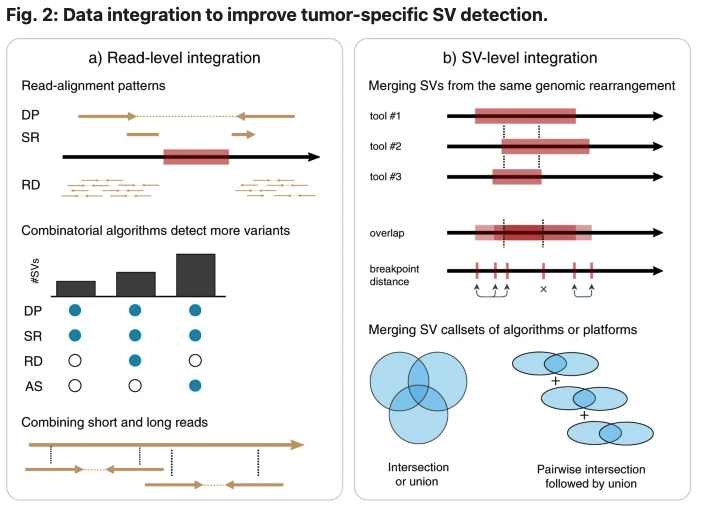
Full-spectrum SV detection with high recall and precision requires integration of multiple algorithms and sequencing technologies to rescue variants that are difficult to resolve through individual methods. Nature Partner Journal Precision Oncology now published an open access review paper on computational challenges in structural variant (SV) detection for precision oncology.
At least 30% of cancers have a known pathogenic SV used in diagnosis or treatment stratification. Classification of tumor-specific SVs is challenging due to inconsistencies in detected breakpoints, derived variant types and biological complexity of some rearrangements. Cancer is generally characterized by acquired genomic aberrations in a broad spectrum of types and sizes, ranging from single nucleotide variants to structural variants (SVs).